Offline tool
An offline tool for identification of targets and prioritizing vaccine candidates from pathogen proteomes, protein-protein interaction network and genome scale metabolic model.
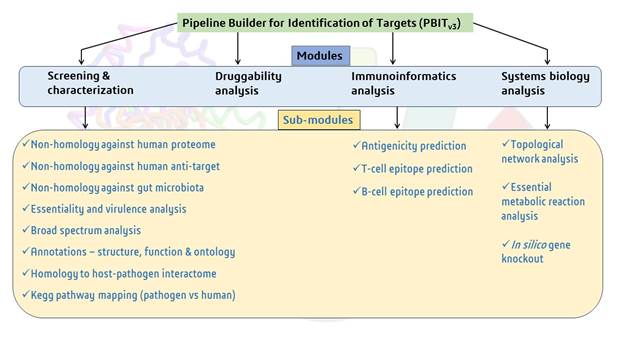
Offline PBIT contains 4 modules and respective submodules:
- Screening & characterization – This module is based on sequence comparison method of pathogen proteins with the human proteome, anti-targets, gut microbiota, essential and virulent proteins and known drug targets, to identify ideal targets. The functionality of each module remains same as that of online PBIT. This module can also be plugged in with the annotation module.
Submodules:
- Non homology analysis against human proteome
- Non homology analysis against human anti-targets
- Non homology analysis against gut microbiota
- Essentiality and virulence analysis
- Broad spectrum analysis
- Annotations – structure, function & ontology
- Homology to human-pathogen interactome
- Kegg pathway mapping (pathogen vs human)
- Druggability - Druggability is the ability of a protein to bind to a drug-like molecule. This module aids to screen druggable targets in the input sequences based on their sequence similarity to known druggable targets. Sequence similarity is computed by BLAST algorithm and the sequence information for druggable targets were obtained from Drugbank database (Version 5.0) containing 4159 non-redundant druggable targets and Therapeutic Target Database (TTD) containing 2,589 targets.
- Immunoinformatics analysis - This module can be used to identify antigenic B-cell and T-cell epitopes of shortlisted pathogen protein sequences.
Submodules:
- Antigenecity Prediction
- T-cell epitope prediction
- B-cell epitope prediction
- Systems biology analysis - Through this module, the protein sequences can be further shortlisted as potential drug targets based on protein-protein interaction network and metabolic model parameters.
Submodules:
- Topological network analysis
- Essential metabolic reaction analysis
- In silico gene knockout
Modules |
Input |
Parameters |
Output |
Screening & Characterization |
FASTA formatted protein sequences |
Choice of submodules
Percent identity
Alignment length cut off
E-value |
FASTA formatted protein sequences |
Druggability Analysis |
FASTA formatted protein sequences |
Percent identity
E-value |
FASTA formatted protein sequences |
Immunoinformatics Analysis |
FASTA formatted protein sequences |
Choice of submodules Window Size |
Excel file containing output of selected submodules |
Systems biology Analysis |
Metabolic model in JSON format
Edge list in CSV format (Network Analysis)
Gene list from model (optional)
|
Choice of submodules
Reaction ids & bounds (optional) |
Excel file containing output of selected submodules |
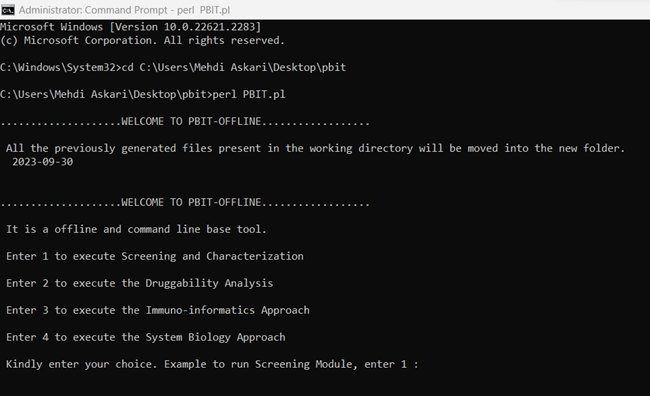
Command line interface for offline PBIT
From Windows command line:
For detailed information on installation and dependencies, please download the manual for PBIT
Further reading:
- Orth, Jeffrey D., Ines Thiele, B. Ø. P. What is flux balance? Nat. Biotechnol. 28, 245–248 (2010).
|
Citation:
Mukherjee S, Kundu I, Askari M, Barai R S, Venkatesh K V, Idicula-Thomas S. Exploring the druggable proteome of Candida species through comprehensive computational analysis. Genomics, Volume 113, Issue 2,2021, Pages 728-739. (https://doi.org/10.1016/j.ygeno.2020.12.040.)
|
|